Pneumonia Classification
Pneumonia an acute disease that is marked by inflammation of lung tissue accompanied by infiltration of alveoli and often bronchioles with white blood cells (such as neutrophils) and fibrinous exudate, is characterized by fever, chills, cough, difficulty in breathing, fatigue, chest pain, and reduced lung expansion, and is typically caused by an infectious agent (such as a bacterium, virus, or fungus). The disease may be classified by where it was acquired, such as community- or hospital-acquired or healthcare-associated pneumonia.
Read file
in this step I’m going to copy file from drive to local google colab bacause if you’re running file from drive it’s going to be much slower compared to local directory in colab.
zip_path = '/content/drive/My\ Drive/BCML/FinalProject/chest_xray_backup.zip'
!cp {zip_path} /content/
!cd /content/
!unzip -q /content/chest_xray_backup.zip -d /content
!rm /content/chest_xray_backup.zip
Importing Necessary Libraries
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import os
import cv2
import datetime
import pandas as pd
from numpy import expand_dims
from sklearn.metrics import classification_report,confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
from sklearn.metrics import auc
import keras
from tensorflow.keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Dense, Conv2D , MaxPool2D , Flatten , Dropout , BatchNormalization,MaxPooling2D
from tensorflow.keras.models import Model
from tensorflow.keras.optimizers import SGD , RMSprop,Adam
from keras.preprocessing.image import ImageDataGenerator,img_to_array, load_img
from keras.callbacks import ReduceLROnPlateau
from tensorflow.keras.callbacks import ModelCheckpoint
from tensorflow.keras.callbacks import TensorBoard
from keras.models import load_model
Definition Function
Code below it’s a function that I used in this project. function get_training_data it’s for splitting data between train, val, and test then give label to data ‘0’ for normal and ‘1’ for pneumonia. compose_dataset is a function for reshaping data. Imagedatagenerator is a function for visualisation image after image augmentation
labels = [ 'NORMAL','PNEUMONIA']
img_size = 200
dataset_dir_train = '/content/chest_xray_backup/train'
dataset_dir_val = '/content/chest_xray_backup/val'
dataset_dir_test = '/content/chest_xray_backup/test'
def get_training_data(data_dir):
data = []
for label in labels:
path = os.path.join(data_dir, label)
class_num = labels.index(label)
for img in os.listdir(path):
try:
img_arr = cv2.imread(os.path.join(path, img), cv2.IMREAD_GRAYSCALE)
resized_arr = cv2.resize(img_arr, (img_size, img_size))
data.append([resized_arr, class_num])
except Exception as e:
print(e)
return np.array(data)
def process_data(img):
img = np.array(img)/255.0
img = np.reshape(img, (200,200,1))
return img
def compose_dataset(df):
data = []
labels = []
for img, label in df.values:
data.append(process_data(img))
labels.append(label)
return np.array(data), np.array(labels)
def imagedatagenerator(param):
test_Image_DG=cv2.imread('/content/chest_xray_backup/train/NORMAL/IM-0115-0001.jpeg', cv2.IMREAD_GRAYSCALE) # Reading an Image
test_Image_DG=cv2.resize(test_Image_DG,(200,200))
test_Image_DG = np.array(test_Image_DG)/255.0
test_Image_DG = np.reshape(test_Image_DG, (200,200,1))
input_image_DG= np.expand_dims(test_Image_DG, axis=0)
if param == "rotation_range":
datagen_rotation_range = ImageDataGenerator(rotation_range =30)
it = datagen_rotation_range.flow(input_image_DG, batch_size=1)
elif param == "zoom_range":
datagen_zoom_range = ImageDataGenerator(zoom_range =0.2)
it = datagen_zoom_range.flow(input_image_DG, batch_size=1)
elif param == "width_shift_range":
datagen_width_shift = ImageDataGenerator(width_shift_range =0.1)
it = datagen_width_shift.flow(input_image_DG, batch_size=1)
elif param == "height_shift_range":
datagen_height_shift_range = ImageDataGenerator(height_shift_range =0.1)
it = datagen_height_shift_range.flow(input_image_DG, batch_size=1)
elif param == "All":
datagen_all = ImageDataGenerator(rotation_range =30,zoom_range =0.2,width_shift_range =0.1,height_shift_range =0.1,horizontal_flip =True)
it = datagen_all.flow(input_image_DG, batch_size=1)
fig=plt.figure(figsize=(30,30))
for i in range(8):
ax=fig.add_subplot(1,8,i+1)
batch = it.next()
image = batch[0]
image = image[:,:,0]
ax.imshow(image,cmap='gray')
return
Get Data Image
train = get_training_data(dataset_dir_train)
test = get_training_data(dataset_dir_test)
val = get_training_data(dataset_dir_val)
Print np array photo
train[0][0]
Output:
array([[61, 68, 75, ..., 26, 12, 0],
[63, 68, 78, ..., 23, 12, 0],
[62, 69, 78, ..., 25, 16, 0],
...,
[ 0, 0, 0, ..., 0, 0, 0],
[ 0, 0, 0, ..., 0, 0, 0],
[ 0, 0, 0, ..., 0, 0, 0]], dtype=uint8)
Print label normal
labels[train[0][1]]
Output:
NORMAL
Print label pneumonia
labels[train[-1][1]]
Output:
PNEUMONIA
Change data to dataframe
train_df = pd.DataFrame(train, columns=['image', 'label'])
test_df = pd.DataFrame(test, columns=['image', 'label'])
val_df = pd.DataFrame(val, columns=['image', 'label'])
print(train_df)
Output:
image label
0 [[61, 68, 75, 76, 84, 89, 89, 93, 91, 88, 86, ... 0
1 [[7, 6, 6, 6, 5, 6, 5, 6, 5, 6, 5, 6, 5, 5, 5,... 0
2 [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,... 0
3 [[78, 78, 75, 139, 169, 137, 120, 116, 117, 12... 0
4 [[33, 26, 32, 37, 44, 50, 53, 52, 58, 60, 63, ... 0
... ... ...
5211 [[31, 29, 29, 31, 31, 34, 34, 31, 29, 28, 28, ... 1
5212 [[16, 15, 20, 21, 21, 254, 23, 19, 255, 20, 21... 1
5213 [[16, 17, 19, 19, 18, 17, 20, 20, 22, 104, 246... 1
5214 [[0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 8, 13, 30, 43,... 1
5215 [[3, 4, 9, 13, 18, 21, 21, 21, 24, 26, 30, 42,... 1
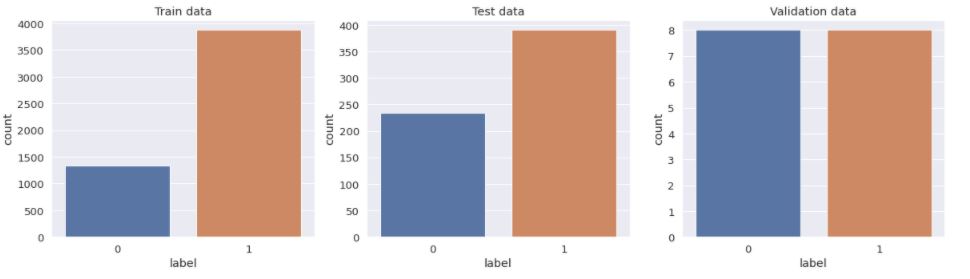
Visualization Data
plt.figure(figsize=(20,5))
plt.subplot(1,3,1)
sns.countplot(train_df['label'])
plt.title('Train data')
plt.subplot(1,3,2)
sns.countplot(test_df['label'])
plt.title('Test data')
plt.subplot(1,3,3)
sns.countplot(val_df['label'])
plt.title('Validation data')
plt.show()
The data seems imbalanced . To increase the no. of training examples, we will use data augmentation
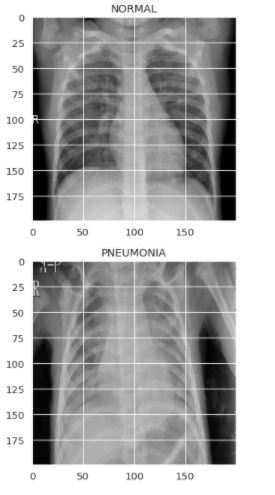
Previewing the images of both the classes
plt.figure(figsize = (5,5))
plt.imshow(train[0][0], cmap='gray')
plt.title(labels[train[0][1]])
plt.figure(figsize = (5,5))
plt.imshow(train[-1][0], cmap='gray')
plt.title(labels[train[-1][1]])
Feature Engineering
We call compose function that we define in a cell before
feature_train, label_train = compose_dataset(train_df)
feature_test, label_test = compose_dataset(test_df)
feature_val, label_val = compose_dataset(val_df)
print('Train data shape: {}, Labels shape: {}'.format(feature_train.shape, label_train.shape))
print('Test data shape: {}, Labels shape: {}'.format(feature_test.shape, label_test.shape))
print('Validation data shape: {}, Labels shape: {}'.format(feature_val.shape, label_val.shape))
we can see the shape it’s changing, its necessary if we want to procces it for CNN.
Data Augmentation
In order to avoid overfitting problem, we need to expand artificially our dataset. We can make your existing dataset even larger. The idea is to alter the training data with small transformations to reproduce the variations. Approaches that alter the training data in ways that change the array representation while keeping the label the same are known as data augmentation techniques. Some popular augmentations people use are grayscales, horizontal flips, vertical flips, random crops, color jitters, translations, rotations, and much more. By applying just a couple of these transformations to our training data, we can easily double or triple the number of training examples and create a very robust model.
train_datagen = ImageDataGenerator(rotation_range = 30, # randomly rotate images in the range (degrees, 0 to 180)
zoom_range = 0.2, # Randomly zoom image
width_shift_range=0.1, # randomly shift images horizontally (fraction of total width)
height_shift_range=0.1 # randomly shift images vertically (fraction of total height)
)
train_datagen.fit(feature_train)
label_train = to_categorical(label_train)
label_test = to_categorical(label_test)
label_val = to_categorical(label_val)
I use to_categorical function from keras to make categorical label.
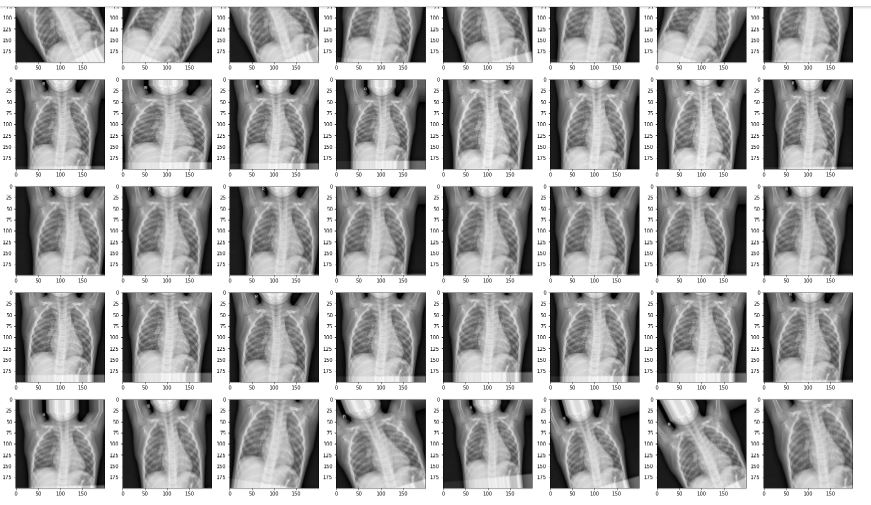
Visualisasi Image Data Generator yang di gunakan
list_param=["rotation_range","zoom_range","width_shift_range","height_shift_range","All"]
for i in list_param:
imagedatagenerator(i)
For the data augmentation, i choosed to :
- Randomly rotate some training images by 30 degrees
- Randomly Zoom by 20% some training images
- Randomly shift images horizontally by 10% of the width
- Randomly shift images vertically by 10% of the height
Once our model is ready, we fit the training dataset.
Model CNN
model = Sequential()
model.add(Conv2D(filters=8, kernel_size=(7,7), padding='same', activation='relu', input_shape=(200, 200, 1)))
model.add(Conv2D(filters=8, kernel_size=(7,7), padding='same', activation='relu'))
model.add(MaxPooling2D(pool_size=(3,3)))
model.add(Conv2D(filters=16, kernel_size=(5,5), padding='same', activation='relu'))
model.add(Conv2D(filters=16, kernel_size=(5,5), padding='same', activation='relu'))
model.add(MaxPooling2D(pool_size=(3,3)))
model.add(Conv2D(filters=32, kernel_size=(3,3), padding='same', activation='relu'))
model.add(Conv2D(filters=32, kernel_size=(3,3), padding='same', activation='relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Conv2D(filters=64, kernel_size=(3,3), padding='same', activation='relu'))
model.add(Conv2D(filters=64, kernel_size=(3,3), padding='same', activation='relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Conv2D(filters=128, kernel_size=(3,3), padding='same', activation='relu'))
model.add(Conv2D(filters=128, kernel_size=(3,3), padding='same', activation='relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.2))
model.add(Dense(2, activation='softmax'))
optimizer = Adam(lr=0.0001, decay=1e-5)
model.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['accuracy'])
model.summary()
I Want to see conv2D layer in my model so I print it for I use next.
i=1
for layer in model.layers:
if 'conv' in layer.name:
filters, bias= layer.get_weights()
print('Filters Shape: '+ str(filters.shape, )+" " + 'Bias Shape: '+str(bias.shape)+ "<---- layer: "+str(i))
print("-----------")
i=i+1
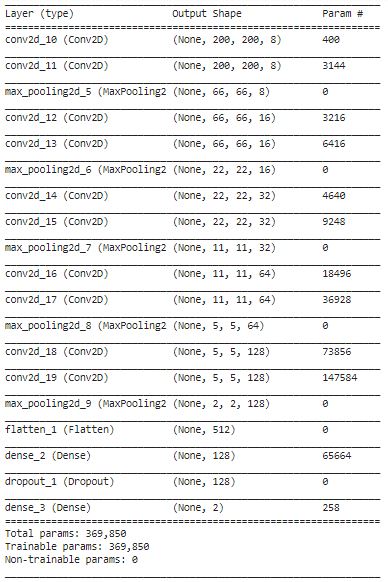
Visualization Layer Filter CNN 1
layer= model.layers
layer_1= layer[0]
filter_1, bias_1= layer_1.get_weights()
print(filter_1.shape, bias_1.shape)
#Normalize the weights
f_min, f_max = filter_1.min(), filter_1.max()
filter_1 = (filter_1 - f_min) / (f_max - f_min)
filter_1 = filter_1[:,:,0]
fig= plt.figure(figsize=(20,20))
for i in range(8):
ax = fig.add_subplot(1,8,i+1)
ax.imshow(filter_1[:,:,i], cmap='gray')
I want to know the filter that cnn use in first conv2d. so i can know about feature map and how cnn works.
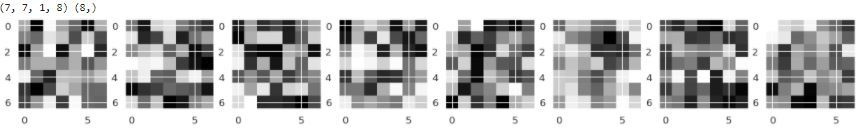
Visualization Layer Filter CNN 2
layer_2= layer[1]
filter_2, bias_2= layer_2.get_weights()
print(filter_2.shape, bias_2.shape)
#Normalize the weights
f_min2, f_max2 = filter_2.min(), filter_2.max()
filter_2 = (filter_2 - f_min2) / (f_max2 - f_min2)
filter_2 = filter_2[:,:,0]
fig= plt.figure(figsize=(20,20))
for i in range(8):
ax = fig.add_subplot(1,8,i+1)
ax.imshow(filter_2[:,:,i], cmap='gray')
as you can see filter in conv2D it’s diffrent although the kernel and neuron it’s a same. this is very interesting because CNN randomly setting a filter for every run model.
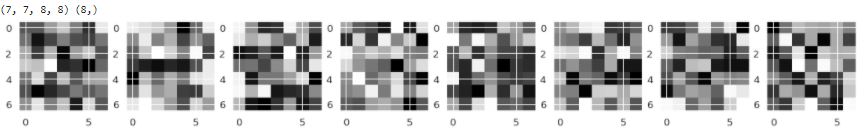
Result of implementation filter CNN 1
inp= model.inputs
out1= model.layers[0].output
feature_map_1= Model(inputs= inp, outputs= out1)
img=cv2.imread('/content/chest_xray_backup/train/NORMAL/IM-0115-0001.jpeg', cv2.IMREAD_GRAYSCALE) # Reading an Image
img=cv2.resize(img,(200,200)) # Resizing an Image
img = np.array(img)/255.0
img = np.reshape(img, (200,200,1))
input_img= np.expand_dims(img, axis=0) # Expanding the dimension
print(input_img.shape) # Printing out the size of the Input Image
#-------------------------------------#---------------------------
f1=feature_map_1.predict(input_img) # predicting out the Image
print(f1.shape) # Let's see the shape
fig= plt.figure(figsize=(20,20))
for i in range(8):
ax=fig.add_subplot(1,8,i+1)
ax.imshow(f1[0,:,:,i],cmap='gray')
now you can see how filter change image in the first Conv2D layer
Result of implementation filter CNN 2
inp= model.inputs
out2= model.layers[1].output
feature_map_2= Model(inputs= inp, outputs= out2)
f2=feature_map_2.predict(input_img) # predicting out the Image
print(f2.shape) # Let's see the shape
fig= plt.figure(figsize=(20,20))
for i in range(8):
ax=fig.add_subplot(1,8,i+1)
ax.imshow(f2[0,:,:,i],cmap='gray')
you can see how filter change image in the second Conv2D layer. now, we can conclude that image change for every conv2d and for every epoch that we define.
Training Model with Validation Set
settings for callbacks
filepath="weights_best.h5"
checkpoint = ModelCheckpoint(filepath, monitor='val_accuracy', verbose=1, save_best_only=True, mode='max')
callbacks_list = [checkpoint]
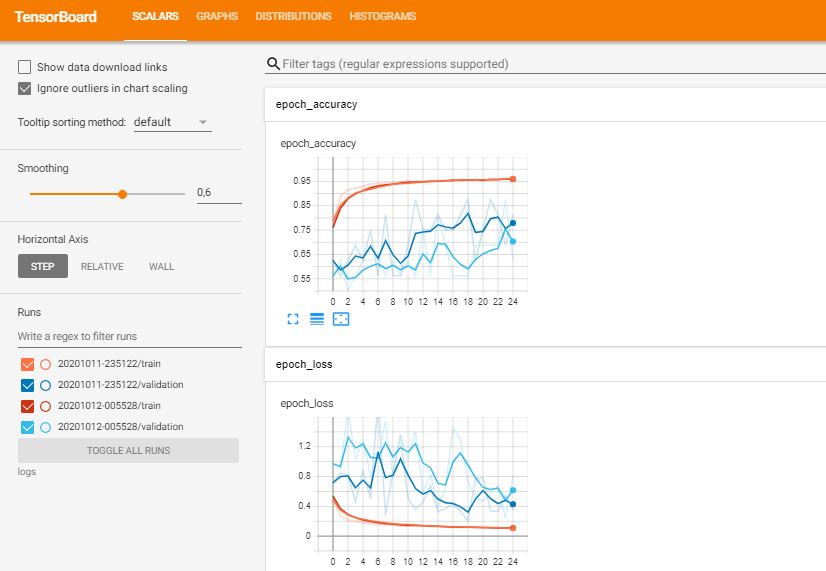
logdir = os.path.join("logs", datetime.datetime.now().strftime("%Y%m%d-%H%M%S")) # Tempat dimana log tensorboard akan di
callbacks_list.append(TensorBoard(logdir, histogram_freq=1))
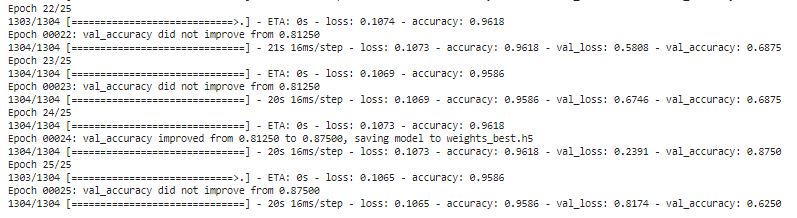
history = model.fit(train_datagen.flow(feature_train,label_train, batch_size=4), validation_data=(feature_val, label_val), epochs = 25, verbose = 1, callbacks=callbacks_list)
%load_ext tensorboard
%tensorboard --logdir logs
Evaluate Model for Testing Datasets
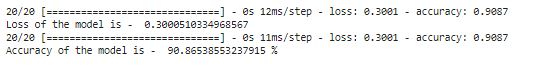
print("Loss of the model is - " , model.evaluate(feature_test,label_test)[0])
print("Accuracy of the model is - " , model.evaluate(feature_test,label_test)[1]*100 , "%")
As you can see we get 90.86% accuracy from Test dataset.
predictions = np.argmax(model.predict(feature_test), axis=-1)
predictions = predictions.reshape(1,-1)[0]
predictions[:100]
label_test_hat = model.predict(feature_test, batch_size=4)
label_test_hat = np.argmax(label_test_hat, axis=1)
label_test = np.argmax(label_test, axis=1)
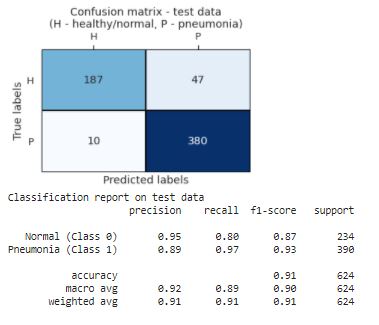
# calculate confusion matrix & classification report
conf_m = confusion_matrix(label_test, label_test_hat)
clas_r = classification_report(label_test, label_test_hat,target_names = ['Normal (Class 0)','Pneumonia (Class 1)'])
# plot confusion matrix as heatmap
plt.figure(figsize=(5,3))
sns.set(font_scale=1.2)
ax = sns.heatmap(conf_m, annot=True,xticklabels=['H', 'P'], yticklabels=['H', 'P'], cbar=False, cmap='Blues',linewidths=1, linecolor='black', fmt='.0f')
plt.yticks(rotation=0)
plt.xlabel('Predicted labels')
plt.ylabel('True labels')
ax.xaxis.set_ticks_position('top')
plt.title('Confusion matrix - test data\n(H - healthy/normal, P - pneumonia)')
plt.show()
# print classification report
print('Classification report on test data')
print(clas_r)
Visualization for Confusion Matrix and Classification Report
Visualization Data Validation
label_val_hat = model.predict(feature_val, batch_size=4)
label_val_hat = np.argmax(label_val_hat, axis=1)
label_val = np.argmax(label_val, axis=1)
plt.figure(figsize=(20,15))
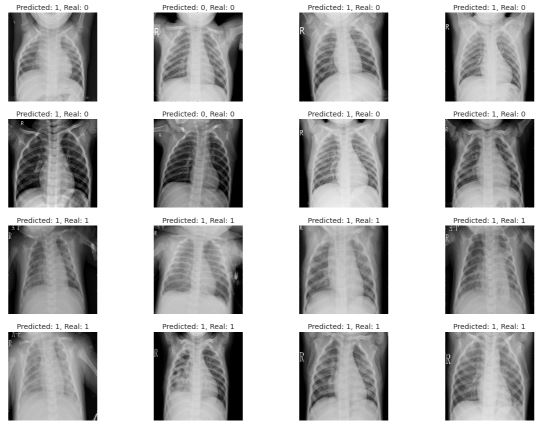
for i,x in enumerate(feature_val):
plt.subplot(4,4,i+1)
plt.imshow(x.reshape(200, 200), cmap='gray')
plt.axis('off')
plt.title('Predicted: {}, Real: {}'.format(label_val_hat[i], label_val[i]))
as you can see in visualization above, how predict works and we can see label before and predict
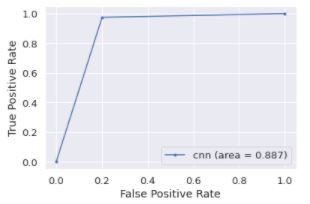
RUC-AUC Curve
AUC - ROC curve is a performance measurement for classification problem at various thresholds settings. ROC is a probability curve and AUC represents degree or measure of separability. It tells how much model is capable of distinguishing between classes.
Data Test
label_pred = model.predict(feature_test,batch_size= 4)
label_pred = np.argmax(label_pred, axis=1)
fpr, tpr, threshold = roc_curve(label_test, label_pred)
auc_test = auc(fpr, tpr)
plt.plot(fpr, tpr, marker='.', label='cnn (area = {:.3f})'.format(auc_test))
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
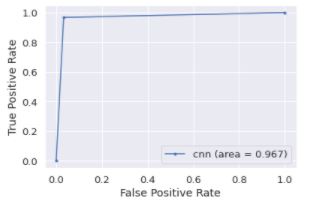
Data Train
train_pred = model.predict(feature_train,batch_size= 4)
train_pred = np.argmax(train_pred, axis=1)
train_label_roc = np.argmax(label_train, axis=1)
fpr_train, tpr_train, threshold = roc_curve(train_label_roc, train_pred)
auc_train = auc(fpr_train, tpr_train)
plt.plot(fpr_train, tpr_train, marker='.', label='cnn (area = {:.3f})'.format(auc_train))
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
Thank you for read my experiment about this dataset. you can also running this program from my heroku web in this link